I am currently a third year PhD student in the Passmore Lab in the MRC Laboratory of Molecular Biology, Cambridge, UK. My research interests lie in using structural biology and in vitro biochemical and biophysical techniques to study macromolecular complexes. In doing so, we can elucidate the mechanisms underlying life’s fundamental processes and hence use this knowledge to understand the basis of disease. I am also a member of Christs College, University of Cambridge.

My current project involves the highly conserved eukaryotic deadenylase Pan2-Pan3. The development and survival of every organism requires gene expression (that is, when/where/how much genes are turned on and off) to be highly regulated. This regulatory process can occur at any stage along the central dogma, from DNA to mRNA to protein. Shortly after transcription, the process by which the DNA template is “read” and “transcribed” into a complementary messenger RNA (mRNA), a polyadenosine (poly(A)) tail is added to the mRNA’s 3′ end. This tail is key in ensuring transcript stability and stimulating translation, the decoding of the mRNA “message” into a functional protein. Once the mature mRNA is exported into the cytoplasm, the poly(A) tail is thought to be bound by a fundamental protein: the poly(A)-binding protein Pab1.
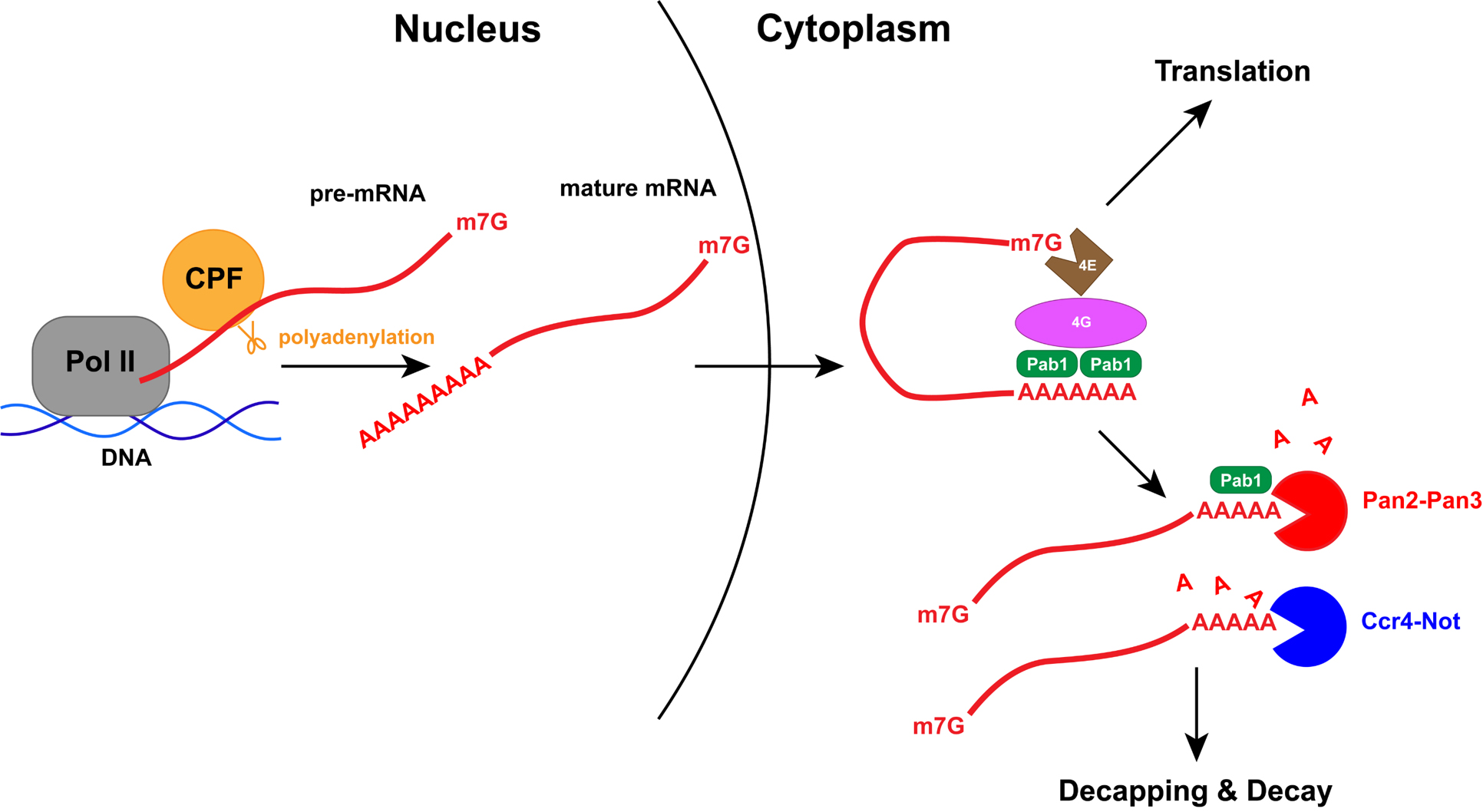
Because of the importance of the poly(A) tail in transcript stability, its dynamic turnover is also highly regulated and thought to be a key step in posttranscriptional regulation of gene expression. In particular, shortening of the poly(A) tail, a process known as deadenylation, is proposed to be the committing and rate-determining step of transcript degradation. Its importance in regulating gene expression is exemplified by developmental diseases when deadenylation is disrupted. In eukaryotes, deadenylation is carried out by two highly conserved but partially redundant multiprotein complexes: Pan2-Pan3 and Ccr4-Not. While the role and mechanism of Ccr4-Not is well-studied, that of Pan2-Pan3 remains unclear.
Using a combination of structural biology and in vitro biochemistry, my current research seeks to understand:
- The physiological role of Pan2-Pan3 and how this distinguishes the complex from Ccr4-Not;
- The mechanistic basis of regulation of Pan2-Pan3 by the poly(A)-binding protein Pab1;
- How Pan2-Pan3 engages with its substrate, the poly(A) tail – thereby understanding the specificity of the Pan2 exonuclease.
(See here).